Blog Uncategorized
The P8 Genetic Variant Affects PYROXD1, a Gene Required for Oxidative Defense
Contents
- Introduction
- Function of PYROXD1
- Evolutionary conservation
- Figure 1. Multiple alignment of protein sequences of PYROXD1 proteins from 248 species of vertebrates shows evolutionary conservation of the amino acid altered by the equine D492H allele.
- Biochemical evidence
- Figure 2. Test of the ability of human PYROXD1 to rescue glr1 yeast cells in the presence of hydrogen peroxide (H2O2).
- Effects of the P8 allele of PYROXD1 in horses
- References
Introduction
The P8 genetic variant that has been part of EquiSeq’s Myopathy Panel since October 2019 is a missense allele of PYROXD1, a gene required for oxidative defense [1]. Mutations of the human PYROXD1 gene are associated with Myofibrillar Myopathy 8 in humans [1]. The P8 allele of equine PYROXD1 changes an aspartic acid (D) to a histidine (H), a nonconservative substitution. Comparison of the amino acid sequence of the PYROXD1 protein across 248 species of vertebrates shows that the position affected by the P8 allele of PYROXD1 is highly conserved across hundreds of millions of years of evolution, with only aspartic acid (D) or asparagine (N) seen in this position. Preliminary biochemical evidence suggests that the P8 allele of PYROXD1 reduces the stability of the protein. There is independent evidence that the basis of Arabian MFM is a defect in oxidative defense affecting the redox state of cysteine thiols.
Function of PYROXD1
PYROXD1 encodes pyridine nucleotide-disulfide oxidoreductase domain 1, an FAD-binding oxidoreductase that uses NAD/NADH as a cofactor to reduce cysteine thiols as part of the cellular oxidative defense.
Mutations in the human PYROXD1 gene are associated with Myofibrillar Myopathy 8 (MFM8), a disease state defined by a breakdown of the integrity of the sarcomeric Z disc and myofibrils, accompanied by the accumulation of aggregates of muscle proteins including desmin, alpha-B-crystallin, and myotilin [1]. Patients show a progressive muscle weakness and atrophy. Association of mutations in PYROXD1 with the disease state in humans is confirmed by experiments in zebrafish, where morpholino knockdowns of the orthologous ryroxD1 gene produce sarcomeric disorganization, aggregates of muscle proteins, and muscle defects that manifest as an impairment of swimming [1].
Evolutionary conservation
The P8 allele, defined by its coordinates in the current genome assembly for horse, EquCab3.0 [2], is chr6:48,924,749 G/C. It is identified in the public SNP database as rs1136260157. There are several models of the PYROXD1 protein that number this position differently. For purposes of this blog post, we will use the protein model XP_001502180.3, which would make this allele D492H.
The equine D492H allele is a nonconservative substitution of a residue that shows evolutionary conservation, as shown in Figure 1.

Figure 1. Multiple alignment of protein sequences of PYROXD1 proteins from 248 species of vertebrates shows evolutionary conservation of the amino acid altered by the equine D492H allele. PYROXD1 protein sequences from 248 species were retrieved from the NCBI nr database using a query sequence consisting of the C-terminal 30 amino acids from the human PYROXD1 protein with the accession ID NP_079130.2 (FENLILNQMNLSSYGEDLLDPNIDIEDYFD). Identical sequences were clustered, producing 48 clusters of sequences, with the largest cluster containing 65 sequences, and 26 sequences being unique. The number of sequences in a cluster are given to the right of each sequence. Each cluster is given a name in the figure. All 248 species names and accession IDs are given in a spreadsheet available for downloading. The 48 sequences were aligned using Clustal Omega [3]. Scores given by Clustal Omega in the next to the last line of the figure are: identical (*), highly conserved (:), conserved (.), and not conserved (). The position of the amino acid affected by the D492H allele is shown in red.
The amino acid affected by the equine D492H allele is highly conserved across vertebrates, with only aspartic acid (D) or asparagine (N) at this position in 248 species including cartilaginous fishes, which are separated from mammals by approximately 450 million years of evolutionary divergence. This does not say that the D492H variant has never occurred in any of the species examined, but rather that whenever it has occurred, it has been cleared from the population by natural selection. This is strong evidence that the D492H allele of PYROXD1 is pathogenic.
Biochemical Evidence
The PYROXD1 gene encodes a thiol reductase with a role in oxidative defense. Glutathione, a tripeptide that contains a cysteine residue, is a key molecule in maintaining the redox state of cysteine thiols. Glutathione exists in two forms, the monomeric reduced form (GSH) and the oxidized dimer (GSSG) in which two molecules of glutathione are covalently joined by a disulfide bond between the cysteine residues. An enzymatic oxidoreductase operates by reducing one molecule while oxidizing another; glutathione is one of the substrates for some of these enzymes, becoming oxidized to the dimer (GSSG) while the other substrate is reduced. Reduced glutathione (GSH) is regenerated by enzymes like glutathione reductase, which uses NADPH as a cofactor.
Yeast cells defective in glutathione reductase (glr1) have an impaired ability to regenerate reduced glutathione (GSH), and hence have an impaired defense against reactive oxygen species like hydrogen peroxide (H2O2). This can be seen in a plate assay where cultures of wild-type yeast cells and cells in which the gene encoding glutathione reductase (glr1) has been deleted are spotted on plates containing 0.2% hydrogen peroxide. This assay shows a clear growth disadvantage for glr1 cells in the presence of hydrogen peroxide [1].
As shown in Figure 2, expression of the human PYROXD1 gene can partially rescue glr1 yeast cells in this assay. Different pathogenic alleles of PYROXD1 identified in human clinical studies have been tested for their ability to reduce the ability of human PYROXD1 to rescue yeast glr1 cells grown in the presence of hydrogen peroxide. The results show that PYROXD1 alleles associated with Myofibrillar Myopathy 8 in human patients reduce the ability of the human PYROXD1 gene to replace yeast glutathione reductase in oxidative defense.

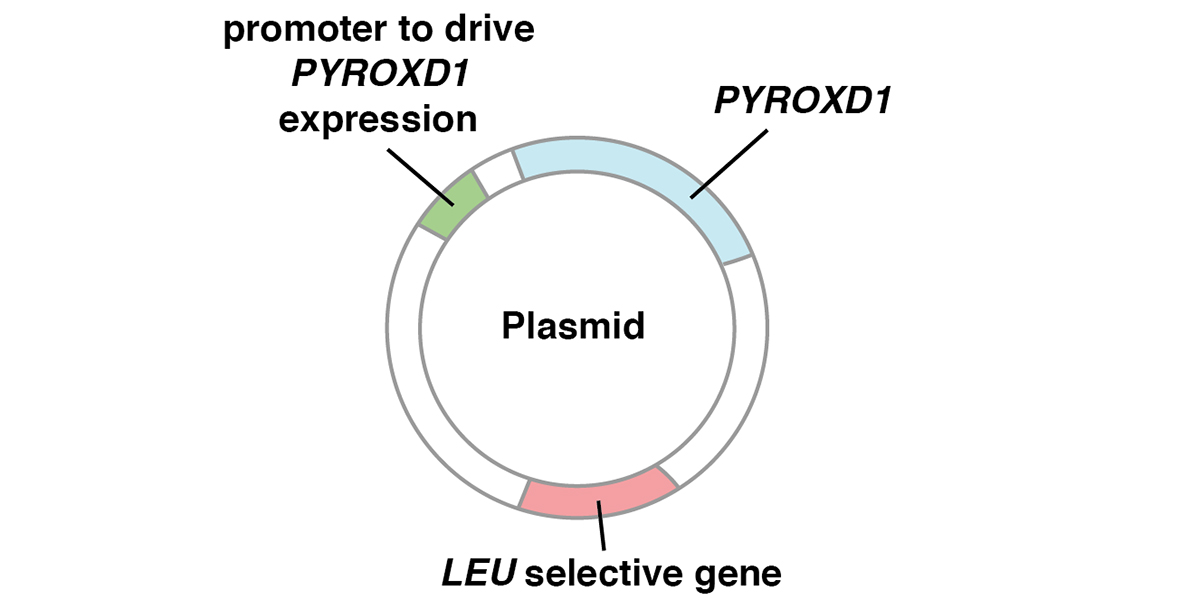
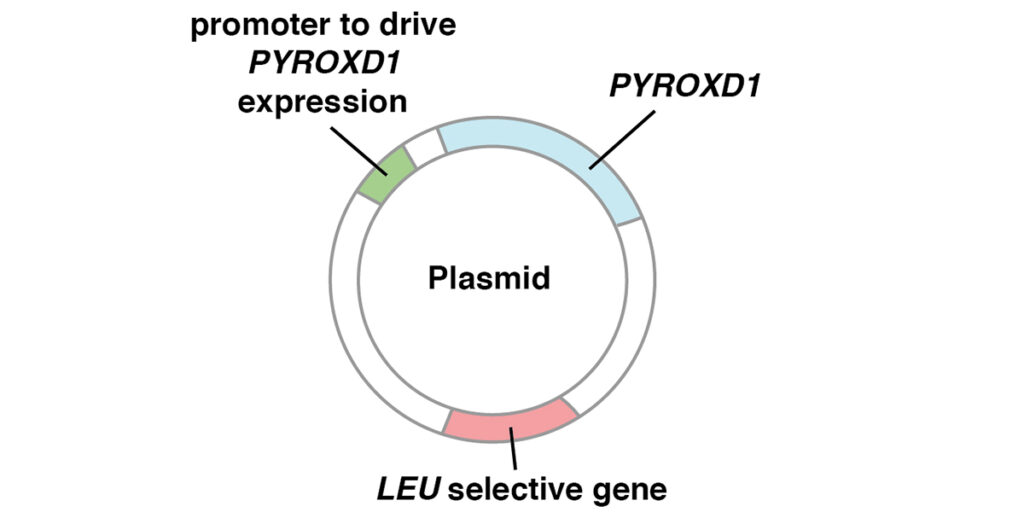
Figure 2. Test of the ability of human PYROXD1 to rescue glr1 yeast cells in the presence of hydrogen peroxide (H2O2). A. Plasmids used to introduce human PYROXD1 into yeast cells. The plasmid, capable of replication in both E. coli and yeast, carries a human PYROXD1 cDNA expressed under the control of a promoter that is active in yeast. The plasmid also contains a LEU selective gene to select for plasmid maintenance in a yeast strain defective in that gene. When cultured in synthetic medium lacking leucine, cells that lose the plasmid are unable to grow. B. Rescue of glr1 yeast cells by human PYROXD1. The first row shows that wild-type yeast cells grow equally well on rich medium or synthetic medium containing hydrogen peroxide (H2O2). The second row shows that cells lacking the yeast glr1 gene encoding glutathione reductase grow poorly in the presence of hydrogen peroxide (H2O2). The third row shows that glr1 cells bearing a plasmid expressing human PYROXD1 are partially rescued. The fourth and fifth row show that the introduction of pathogenic missense alleles (PYROXD1-N155S and PYROXD1-Q372H) reduces the ability of the human PYROXD1 to rescue glr1 cells in the presence of hydrogen peroxide (H2O2). (Adapted from [1]).
Introduction of the equine D492H allele into the human PYROXD1 gene similarly reduces the ability of the human PYROXD1 gene to replace yeast glutathione reductase in oxidative defense (not shown). In addition, Western blot analysis shows that in contrast to human pathogenic missense alleles, the D492H allele reduces the steady-state level of the human PYROXD1 protein in yeast cells. This suggests that the D492H allele decreases that half-life of the protein, possibly by inducing misfolding.
The equine PYROXD1 gene also rescues glr1 yeast cells grown in the presence of hydrogen peroxide. Introduction of the D492H allele into the equine gene reduces the ability of the PYROXD1 protein to replace yeast glutathione reductase in oxidative defense (not shown).
These preliminary results (not yet published in a peer-reviewed scientific journal) demonstrate that the P8 allele of PYROXD1 reduces the function of gene known to be dosage sensitive [4]. The P8 allele of PYROXD1 is therefore expected to be pathogenic in horses.
Effects of the P8 allele of PYROXD1 in horses
A targeted mutation of PYROXD1 in mouse is a recessive lethal and has effects in heterozygotes, showing that PYROXD1 is dosage sensitive (haploinsufficient) [4]. Our initial survey of the effects of the equine P8 allele suggest that it is semidominant and interacts with other mutations associated with Polysaccharide Storage Myopathy type 2 (PSSM2) and Myofibrillar Myopathy (MFM). Homozygotes (P8/P8) are more strongly affected.
The P8 allele of PYROXD1 is widespread among horse breeds. We have detected the P8 allele in Icelandics, Miniatures, Arabians, Thoroughbreds, Warmbloods, and stock breeds (Quarter Horse, Paints, Appaloosas, and POAs). A recent publication identifies the P8 variant of PYROXD1 in two uncharacterized Quarter Horses among 28 horses of various breeds tested. The P8 variant was not seen in any of the eight Warmbloods diagnosed with MFM by muscle biopsy or eight Warmbloods scored as normal by muscle biopsy [5].
A form of Myofibrillar Myopathy (MFM) seen in Arabians is characterized by symptoms of exercise intolerance, breakdown of the Z disc and myofibrils, and accumulation of muscle protein aggregates [6, 7]. Expression profiling and proteomic analysis of Arabians with this form of MFM suggest a defect in oxidative defense, specifically in the redox state of cysteine thiols [7]. The authors propose that a deficiency of cysteine-containing antioxidants is responsible for the aggregation of muscle proteins seen in this disease state.
A survey of 316 Arabians recruited from Arabian interest groups on Facebook without respect to symptoms gives an allele frequency of P8 about 0.25, which means that the population of Arabians is about 6% P8/P8, 37% n/P8, and 57% n/n. The high incidence of the P8 allele of PYROXD1 in Arabians suggest that this mutation may be part of the basis of Arabian MFM, although the horses in the study cited above [7] were not genotyped for this variant.
References
[1] O’Grady et al. (2016). Variants in the oxidoreductase PYROXD1 cause early-onset myopathy with internalized nuclei and myofibrillar disorganization. Am J Hum Genet 99(5): 1086-1105. PMID: 27745833
[2] Kalbfleisch TS et al. (2008). Improved reference genome for the domestic horse increases assembly contiguity and composition. Commun Biol. 1: 197. PMID: 30456315
[3] Madeira F et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res, 47(W1): W636–W641. PMID: 30976793
[4] Dickinson ME et al. (2016). High-throughput discovery of novel developmental phenotypes. Nature 537(7621): 508-514. PMID: 27626380
[5] Williams ZJ et al. (2020). Candidate gene expression and coding sequence variants in Warmblood horses with myofibrillar myopathy. Equine Vet J. 00: 1-10. PMID: 32453872
[6] Valberg, SJ et al. (2016). Suspected myofibrillar myopathy in Arabian horses with a history of exertional rhabdomyolysis. Equine Vet J. 48(5): 548-56. PMID: 26234161
[7] Valberg, SJ et al. (2018). Proteome and transcriptome profiling of equine myofibrillar myopathy identifies diminished peroxiredoxin 6 and altered cysteine metabolic pathways. Physiol Genomics. 50(12): 1036-1050. PMID: 30289745
Share this post
From the blog
The latest industry news, interviews, technologies, and resources.
Experienced Equestrian Joins EquiSeq Board
Albuquerque, New Mexico, USA June 20, 2025 EquiSeq, a leader in developing genetic…