Our blog
Resources and insights
The latest industry news, interviews, technologies, and resources.
Blog categories
Blog
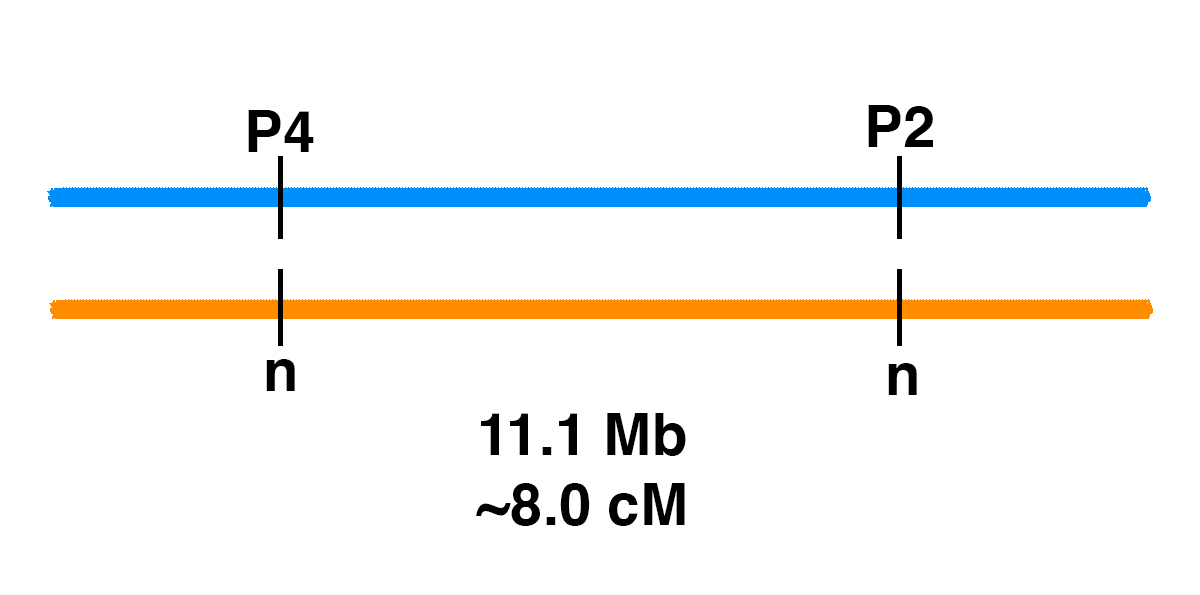
Genetic Linkage of P2 and P4
How can you figure out the chances of different genotypes when breeding horses?Monohybrid CrossIt’s easy for one gene. Let’s say that there is a stallion that has one copy of one of the genetic variants associated with PSSM2, the P2 variant (MYOT-S232P). This stallion is heterozygous (n/P2), meaning that he has one normal copy of the MYOT gene and one copy with the P2 mutation.If the stallion is bred to a mare that is clear (n/n) for the P2 allele, the Punnett square below shows the odds.Gametes (sperm and eggs) contain a single copy of each gene in the horse. The stallion produces two kinds of sperm in equal frequency: sperm with P2, and sperm with the normal allele (n). The mare produces eggs with one of the two normal alleles (n).This means that there is a 50% chance of a foal that is n/P2, and a 50% chance of a foal that is n/n.Dihybrid CrossWhat if we are following two genes? In general, two gene pairs will show independent assortment. This means that the segregation of one pair of alleles into gametes will not influence the segregation of another pair of alleles.If we tossed two coins at the same…
Blog
Filamin C (FLNC)
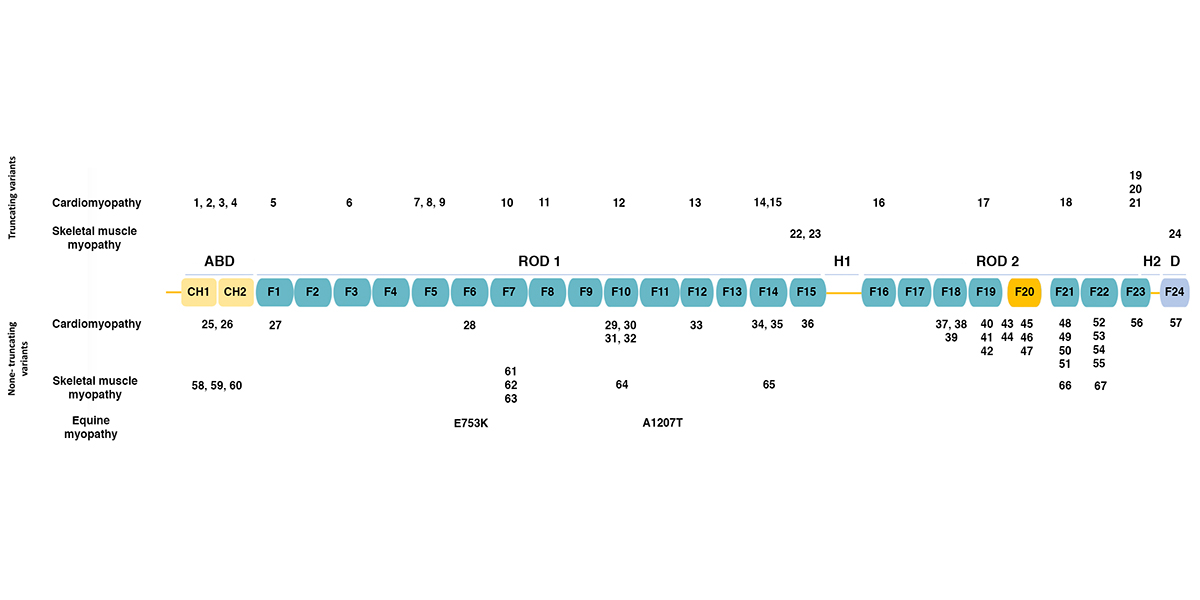
Filamins are a family of actin-binding proteins. In humans and other mammals, there are three filamins: filamin A, filamin B, and filamin C. Filamins A and B are widely expressed, while filamin C expression appears limited to skeletal and cardiac muscle. The figure below (adapted from [1]) shows the domain structure of filamin C. Filamins have an amino-terminal actin-binding domain consisting of two calponin-homology (CH) domains. The rest of the protein consists of 24 immunoglobulin-like filamin domains with two hinge domains. Filamin C differs from filamins A and B in a novel domain 20 that interacts with Z disc proteins, including myotilin and myozenins. While filamin domains 1-15 form a rod, domain 20 is folded over domains 19 and 21 [2]. Filamin domain 24 is required for homodimerization. The figure also shows the positions of human mutations associated with various types of cardiomyopathy and skeletal muscle myopathy, as summarized in the table below. Human FLNC Alleles Associated with Cardiomyopathy Figurea Alleleb Domainc Mutation Typed Citation 1 Y7Tfs*51 ABD frameshift 1 2 Y83* ABD termination 3 25 F106L ABD missense 4 3 E108* ABD termination 5 26 V123A ABD missense 5 4 E238Rfs*14 ABD frameshift 1 5 R269* Filamin 1 termination…
Blog
P3 Allele of FLNC is Associated with Myofibrillar Myopathy in Horses
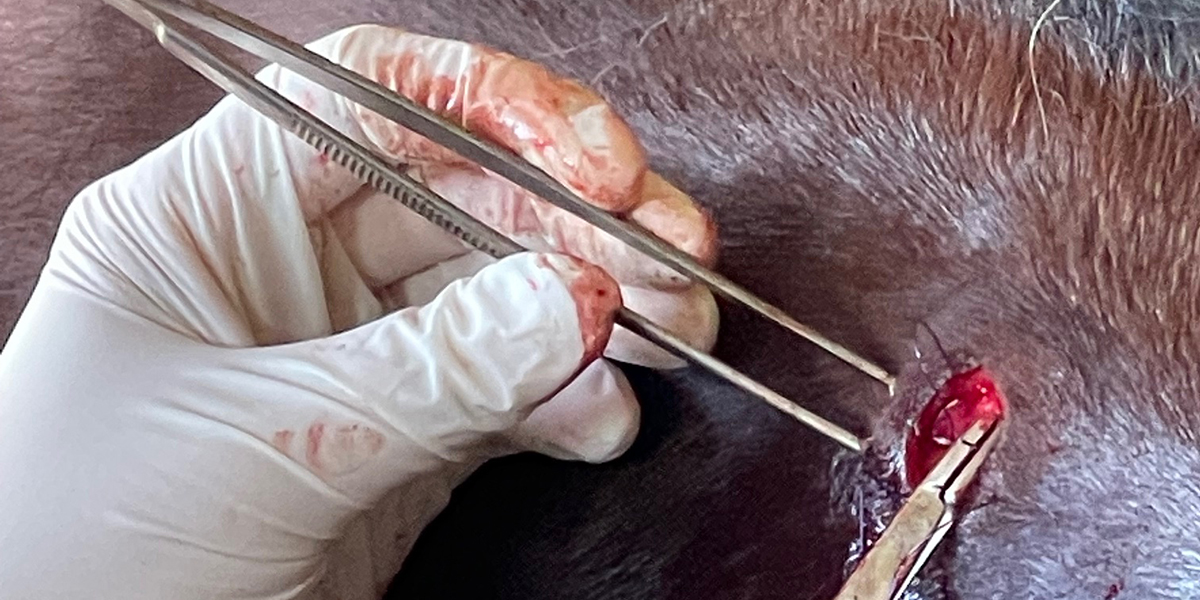
In human patients, mutations in the gene encoding filamin C (FLNC) are associated with Myofibrillar Myopathy 5 [1]. In a prior blog post, we reviewed the human mutations in FLNC associated with the disease state Myofibrillar Myopathy 5 (MFM5) and various cardiomyopathies. We also presented the specific changes associated with the equine P3 allele of FLNC, a haplotype that bears two different missense alleles of FLNC: E753K in Ig-like domain 6 and A1270T in Ig-like domain 11. In this blog post, we review bioinformatic evidence associating the P3 allele of FLNC with Myofibrillar Myopathy in horses. The association of the P3 allele of FLNC with Myofibrillar Myopathy in horses was first made in a horse diagnosed with Myofibrillar Myopathy (MFM) through the identification of desmin-positive inclusions observed in muscle tissue. The horse displayed symptoms of exercise intolerance. A genetic test for GYS1-R309H (P1) ruled out Polysaccharide Storage Myopathy type 1 (PSSM1). The desmin staining technique was relatively new at the time of the muscle biopsy on this horse; the presence of desmin-positive inclusions was diagnostic of MFM in distinction to other types of Polysaccharide Storage Myopathy type 2 (PSSM2). The association of the P3 allele of FLNC with Myofibrillar Myopathy…
Blog
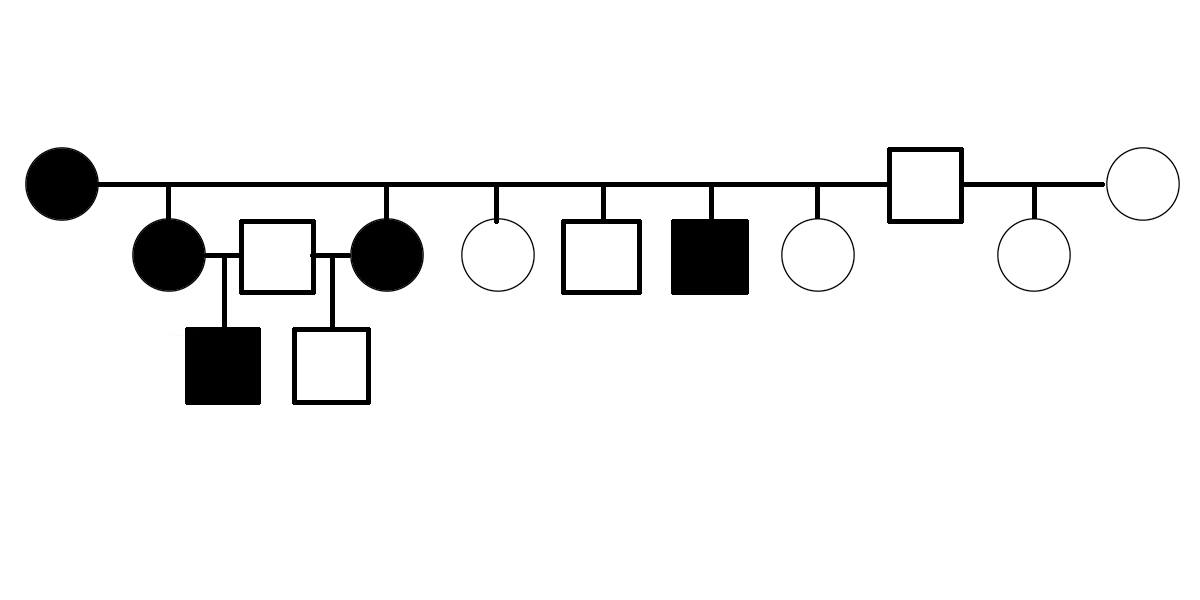
Is PSSM2 Genetic?
Is PSSM2 genetic? By genetic, we mean that it is an inherited condition, transmitted from parents to offspring rather than resulting exclusively from environmental conditions, such as diet or exposure to toxins or pathogens. What do we mean by PSSM2? PSSM2 is an abbreviation for Polysaccharide Storage Myopathy type 2 [1]. It is a disease state in horses initially defined by the following criteria: symptoms of exercise intolerance, a negative DNA test for GYS1-R309H (a genetic variant of the gene encoding glycogen synthase that is associated with the disease state PSSM1), and abnormalities observed in muscle biopsy [1]. The first evidence suggesting that PSSM2 is genetic was presented in 2007, when Dr. Molly McCue presented a pedigree of affected Quarter Horses related by descent from a common ancestor in her PhD dissertation. Related horses were diagnosed with PSSM2 by muscle biopsy. Polysaccharide Storage Myopathy type 1 (PSSM1) was shown to be associated with GYS1-R309H in 2008 [2]. The GYS1-R309H variant has been shown to encode a constitutively activated glycogen synthase [3]. The improper regulation of this enzyme pushes the equilibrium between glycogen synthesis and degradation in the direction of glycogen synthesis, even when the affected horse’s energy needs would normally…
Blog
Myotilin
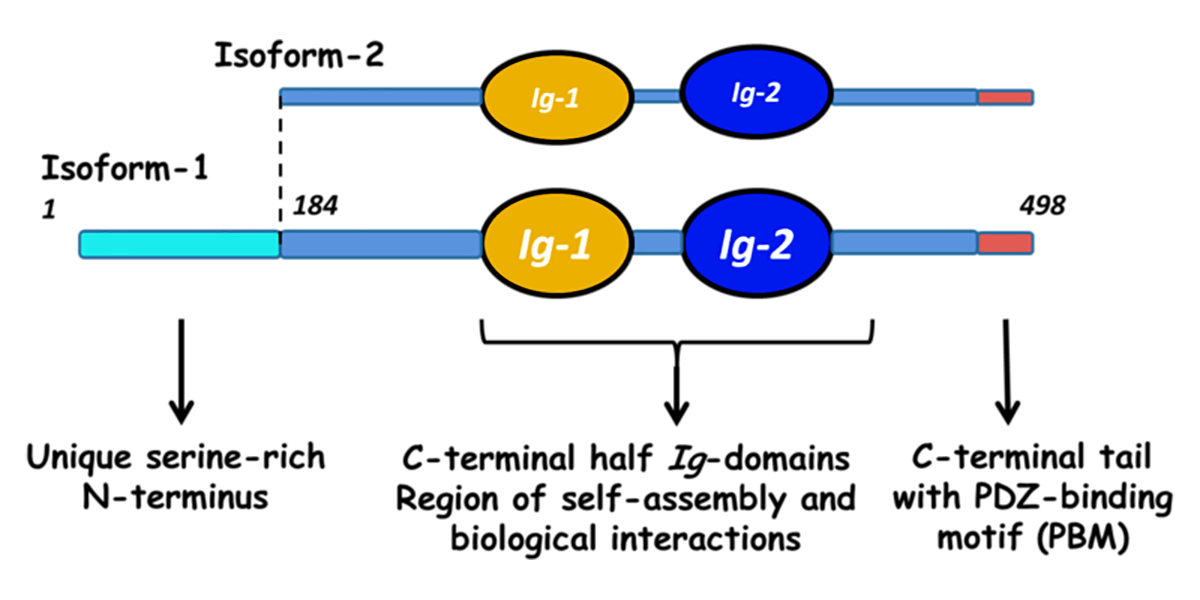
Myotilin is a muscle protein encoded by the MYOT gene. Myotilin is a structural protein of muscle that localizes to the Z discs. Myotilin contains two Immunoglobulin-like (Ig-like) domains similar to those of palladin, myopalladin, titin, and other proteins. The amino-terminal portion of myotilin contains a large number of serine residues and is not similar to any other known protein. Human patients with Limb-Girdle Muscular Dystrophy (LGMD) or Myofibrillar Myopathy (MFM) have been found to have mutations in MYOT. Both LGMD and MFM are disease states associated with mutations in a number of different genes. A substantial fraction of human patients with LGMD or MFM do not receive a molecular diagnosis, suggesting that there are undiscovered genetic variants responsible for some of these cases. Human patients with LGMD that have mutations in MYOT are diagnosed with Limb-Girdle Muscular Dystrophy 1A (LGMD1A), while human patients with MFM that have mutations in MYOT are diagnosed with Myofibrillar Myopathy 3 (MFM3). Pathogenic mutations in MYOT are dominant missense alleles. Known pathogenic alleles include S55F, T57I, S60C, S60F, and S95I [1, 2, 3]. There is a substantial difference in the age of onset and the symptoms of human patients with MYOT mutations, even among…
Blog
Arabian MFM
This was written in response to a post on the PSSM Forum on Facebook. Negative nitrogen balance is a normal process that is helpful under the right circumstances. It is not confined to PSSM horses. You have certainly experienced it yourself. There are some circumstances under which you cannot consume enough dietary protein to meet your needs for amino acids. When you have the flu, for example, your immune system is working hard and you need a lot of amino acids to synthesize proteins. Your body tears down muscle in order break protein down into amino acids for use elsewhere. Some of the amino acids are burned for energy. Skeletal or striated muscle is made up of basic units called sarcomeres (part b of the image above shows the structure of a sarcomere). The sarcomere is bounded by Z discs. Attached to the Z disc are thin filaments composed of actin that overlap thick filaments composed of myosin in the center of the sarcomere. When a muscle contracts, the thick filaments slide relative to the thin filaments. Because the thin filaments are anchored to the Z discs, this sliding causes the sarcomere to contract. Myofibrillar Myopathy is a term borrowed from human…
Blog
Polysaccharide Storage Myopathy type 2 (PSSM2)
Polysaccharide Storage Myopathy type 2 (PSSM2) is a type of exercise intolerance seen in horses. Polysaccharide Storage Myopathy type 2 (PSSM2) is a disease state defined by symptoms of exercise intolerance, absence of the genetic variant GYS1-R309H (P1) that is associated with Polysaccharide Storage Myopathy type 1 (PSSM1) [1], and abnormalities observed in muscle biopsy. The PSSM2 disease state was originally defined during investigation of PSSM1, when it was discovered that some horses with exercise intolerance test negative for GYS1-R309H (P1) and lack the enlarged, amylase-resistant glycogen granules seen in muscle biopsies from horses with the GYS1-R309H (P1) variant [2]. Despite a decade of research since PSSM2 was first described, there is no evidence that PSSM2 is a disorder of carbohydrate metabolism. PSSM2 was originally postulated to be a disorder of glycogen metabolism because muscle biopsies of horses with PSSM2 showed clumping of glycogen granules of normal size. A study of Arabian Myofibrillar Myopathy (MFM) showed that the clumping of glycogen granules in regions of myofibrillar disorganization gives the false appearance of a glycogen storage disorder [3]. Several research teams (including EquiSeq) have performed whole genome sequencing on horses diagnosed with PSSM2 and have failed to find any genetic variants…
Blog
Polysaccharide Storage Myopathy type 1 (PSSM1)
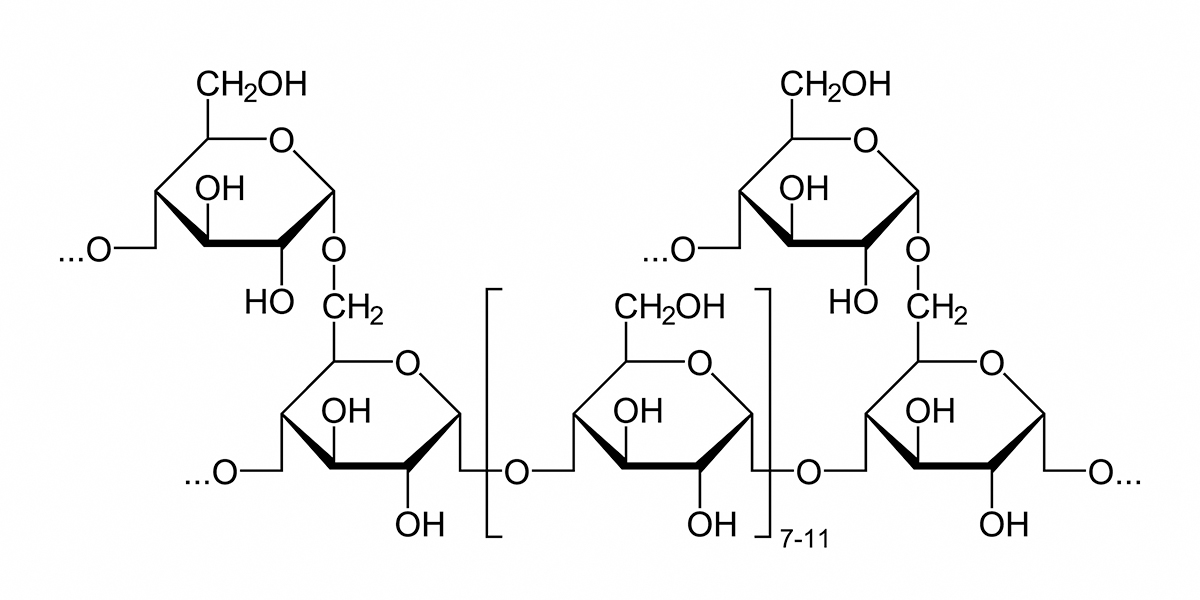
Polysaccharide Storage Myopathy type 1 (PSSM1) is a disease state in horses caused by an inherited defect in glycogen metabolism. Polysaccharide Storage Myopathy type 1 (PSSM1) is a form of exercise intolerance. Horses with Polysaccharide Storage Myopathy type 1 (PSSM1) exhibit exercise-induced symptoms including reluctance to move, pain, stiffness, tremors, and profuse sweating. Serum creatine kinase (CK) and aspartate aminotransferase (AST) are elevated, indicating muscle damage. In severe episodes, myoglobinuria (coffee-colored urine due to the presence of myoglobin), inability to stand, and death can occur. Glycogen is a complex polymer of glucose that serves as a store of glucose in liver and skeletal muscle. As shown in the image below, a glycogen granule consists of multiple branched chains of glucose units. Most of the glucose units are joined by a-1,4 linkages, with a-1,6 linkages at the branch points. The synthesis and breakdown of glycogen are controlled by different enzymes. Glycogen synthase and the branching enzyme synthesize glycogen, while glycogen phosphorylase and the debranching enzyme break it down. Glycogen synthesis and breakdown is regulated in response to energy needs and the availability of glucose. Muscle biopsy of horses with PSSM1 reveals the presence of enlarged glycogen granules that are amylase-resistant. During…
Blog
Jax
Jax is n/P1, n/P2, n/Px. I got him when he was an unbroke 4 year old, and he was quirky that first year but then had over 2 great years of riding. He didn’t go highly symptomatic until he was 8 years old, but right as he turned 8 he crashed hard. Early symptoms included shaky on trailer rides, sticky stifles when out of work, and on/off lameness. As he became more symptomatic he began urinating small amounts of dark brown at the end of the stream – he did this for about a year before major symptoms, right around the time he became overly spooky and started bolting under saddle. These symptoms progressed to shakiness verging on spasms during trailer rides (has gone into full-blown tie up on longer trailer rides), sticky stifles at all times to the point he could barely trot, lumbar and skin pain that would cause him to drop from pressure, couldn’t hold up legs for farrier, and more drastic behavioral changes (aggression towards pasture mates, rearing during handwalks, crow hopping under saddle – but no full-on bucking, etc.). He started twisting his hind legs when walking, had a strange hitching gait in the back…
Blog
Negative Nitrogen Balance
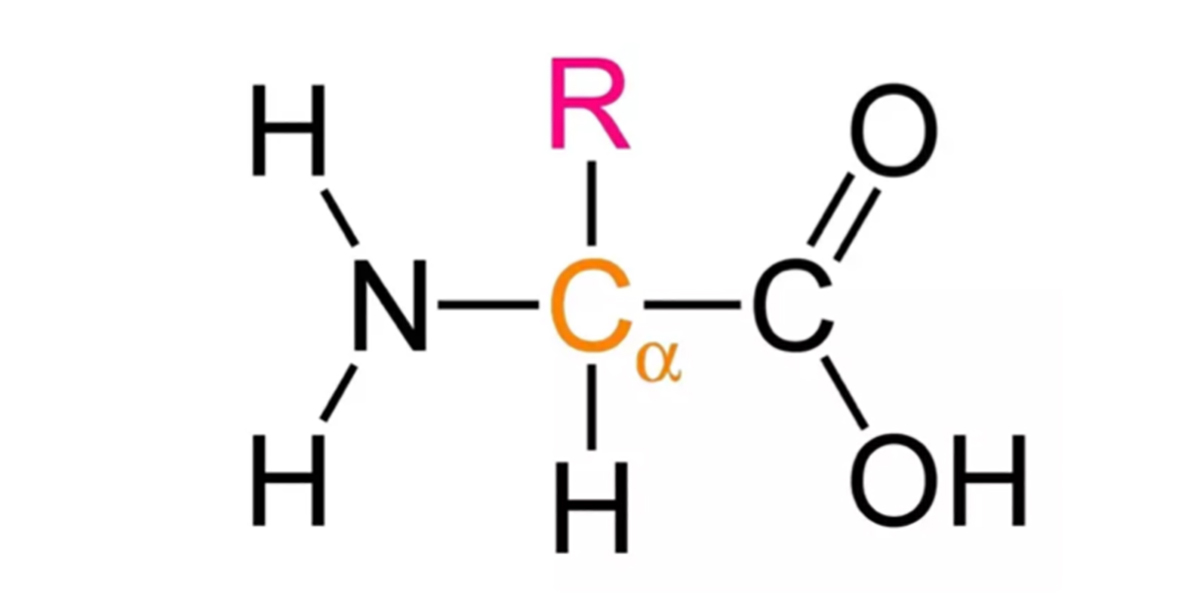
This was written in response to a post on the PSSM Forum on Facebook. Negative nitrogen balance is a normal process that is helpful under the right circumstances. It is not confined to PSSM horses. You have certainly experienced it yourself. There are some circumstances under which you cannot consume enough dietary protein to meet your needs for amino acids. When you have the flu, for example, your immune system is working hard and you need a lot of amino acids to synthesize proteins. Your body tears down muscle in order break protein down into amino acids for use elsewhere. Some of the amino acids are burned for energy. All amino acids have a chemical structure with a central carbon atom bonded to four different parts: an amino group, a carboxyl group, a hydrogen atom, and an “R” group that is a different group for each amino acid. When an amino acid is burned for energy, one of the first steps is the removal of the amino group. If this is not used to synthesize one of the nonessential amino acids, it is discarded as waste. To do that, it enters a metabolic cycle called the urea cycle and is…
- « Previous
- 1
- 2