News
EQUISEQ DISCLOSES GENES AFFECTED BY P2, P3, AND P4 GENETIC VARIANTS
Albuquerque, NM
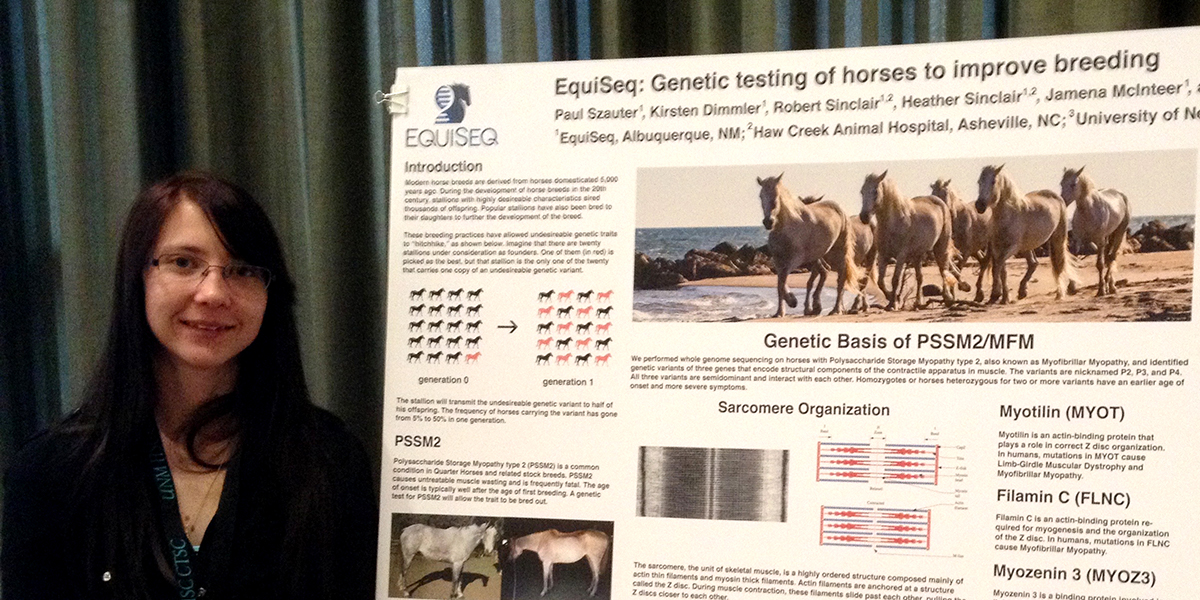
EquiSeq today disclosed the genes affected by the P2, P3, and P4 genetic variants that are part of its Myopathy Panel. The genetic variants are in part responsible for Polysaccharide Storage Myopathy type 2 (PSSM2), also known as Myofibrillar Myopathy (MFM). The genes affected by the P2, P3, and P4 genetic variants were disclosed at a poster presentation at the 2018 BioVenture Partnership Event at the University of New Mexico on March 7.
The P2 genetic variant affects MYOT, the gene encoding myotilin. The P2 variant is a missense allele that causes a nonconservative amino acid substitution in a highly conserved position in the myotilin protein. Myotilin is an actin-binding protein that is part of the Z disc, the part of the sarcomere to which actin thin filaments are anchored. Mutations in human myotilin are the cause of Myofibrillar Myopathy 3 and Limb-Girdle Muscular Dystrophy 1A.
The P3 genetic variant affects FLNC, the gene encoding filamin C. The P3 genetic variant is a pair of missense alleles that cause two nonconservative amino acid substitutions in highly conserved positions in the filamin C protein. Filamin C is a muscle-specific member of the filamin family, an actin-binding protein that is also part of the Z disc. Mutations in human filamin C are the cause of Myofibrillar Myopathy 5 and two forms of hereditary cardiomyopathy.
The P4 genetic variant affects MYOZ3, the gene encoding myozenin 3. The P4 genetic variant is a missense allele that causes a nonconservative amino acid substitution in a highly conserved position in the myozenin 3 protein. Myozenin 3 is a component of the Z disc that binds other Z disc proteins. It has been shown to be important for myogenesis in cultured mouse cells, but pathogenic variants in human have not yet been identified. About half of the cases of human Myofibrillar Myopathy cannot be assigned to any of the eight known genes; it is possible that variants of human MYOZ3 are the cause of some cases.
EquiSeq has not yet disclosed the specific amino acid substitutions caused by these genetic variants, and has also not disclosed any of the supporting evidence that these variants are pathogenic. These data will be presented in a future peer-reviewed scientific publication.
It is clear that there are other genetic variants associated with equine myopathy. EquiSeq’s research team is currently evaluating several newly-discovered genetic variants that may become part of its Myopathy Panel in the future.
All questions should be directed to EquiSeq’s Chief Scientific Officer, Paul Szauter, Ph.D. (pszauter@gmail.com).
Share this post
From the blog
The latest industry news, interviews, technologies, and resources.
Study of horse genomes explores genetic burden
A team of researchers at the University of Minnesota and the University of...
What is PSSM?
Polysaccharide Storage Myopathy (PSSM) is a form of equine exercise intolerance characterized by…